How MERFISH 2.0 Works
Multiplexed Error-Robust Fluorescence in situ Hybridization
Reveal deeper layers of biological complexity with MERFISH 2.0, a single-molecule FISH-based imaging technology for spatially resolved transcriptomics, where optimized chemistry drives sharper resolution for mapping transcripts and cell types with greater precision.
MERFISH achieves unmatched multiplexing and subcellular resolution by assigning each RNA transcript a unique binary barcode decoded through iterative imaging rounds for accurate, reproducible results. Optimized probe chemistry and enhanced readout design boost sensitivity and specificity, even in degraded or archival FFPE samples, expanding transcript detection across diverse tissue types.
By localizing transcripts with nanometer precision, MERFISH 2.0 reveals the spatial organization of genes, cell states, and interactions, creating molecular maps that link expression to function.
- Combinatorial Labeling
- Sequential Imaging
- Error Robust Barcoding

- Perfected RNA Anchoring – Maintains transcript integrity and probe accessibility.
- Enhanced Probe Binding – Maximizes occupancy at target sites for improved coverage.
- Amplified Readout Probes – Boosts fluorescent signal for higher sensitivity and accuracy.
MERFISH 2.0 is Built Upon MERFISH Chemistry
Map Challenging Tissues
Acquire high-quality, reproducible spatial data from even the most difficult samples, including archival FFPE and fresh-frozen tissues.
High Dynamic Range Transcript Capture
Improved sensitivity detects more RNA molecules with uncompromised specificity and subcellular resolution, revealing subtle expression gradients.
More Cells, More Insights
Generate richer datasets with more cells passing quality filters, enabling higher transcript detection and deeper biological context per sample.
Identify Cell Types and States
Distinguish rare and abundant cell populations across complex tissues to illuminate the mechanisms that drive health and disease.
Uncover More Spatial Interactions
Explore molecular and cellular crosstalk within intact tissue architecture to inform disease models and therapeutic discovery.
Proven, Scalable MERFISH Chemistry
Built on decades of MERFISH spatially resolved transcriptomics innovation, MERFISH 2.0 delivers scalable, high-confidence results.

Why Choose MERFISH 2.0?
With MERFISH 2.0, researchers can:
- Map cell types and spatial relationships with unmatched precision
- Detect transcriptomic variations to study cell states and disease mechanisms
- Leverage archival FFPE samples for high-quality data and impactful discoveries
- Customize experiments to fit evolving research questions
More transcripts. More cells. More insights.
Peer-Reviewed Publications
Trusted by researchers worldwide, MERFISH is supported by 100s of peer-reviewed studies showcasing spatially resolved transcriptomics to decode tissue complexity and cellular organization.
What Customers Are Saying

Marco Genua, Giulia Chianella, Antonella Santoro, San Raffaele-Telethon Institute for Gene Therapy (SR-TIGET)

Marco Genua, San Raffaele-Telethon Institute for Gene Therapy (SR-TIGET)

Raymond Yip, WEHI, Australia

University Hospital Tübingen, Germany
Explore MERFISH Data
Access datasets from MERFISH 2.0 spatially resolved transcriptomics experiments and legacy MERFISH chemistry, showcasing the system’s sensitivity across diverse tissues.
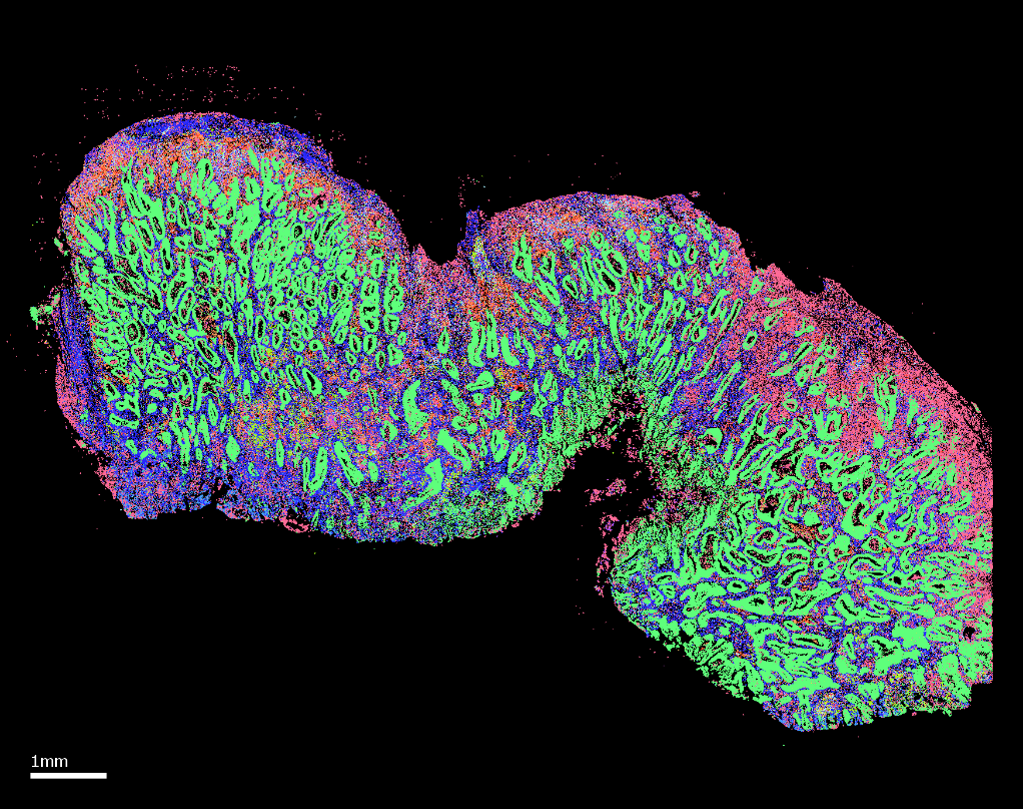
MERFISH 2.0
From Breast and Colon Cancer to Human and Mouse Brain, this data release showcases how MERFISH 2.0 pushes the boundaries of spatial transcriptomics.
ACCESS THE DATA SET
MERSCOPE Ultra Data Release
Explore the spatial transcriptome with the latest generation of the MERSCOPE platform.
ACCESS THE DATA SET
Vizgen MERFISH (1.0) Mouse Receptor Map
Full coronal brain slices highlighting GPCR and RTK expression using legacy MERFISH chemistry.
ACCESS THE DATA SET