
MERSCOPE Ultraᵀᴹ
Map transcripts across 3 cm2 of tissue on a single slide with MERSCOPE Ultra™, a high-resolution in situ spatial imaging platform that offers a tissue-wide view of up to 1,000 custom genes at single-cell resolution.

Why MERSCOPE Ultra?
Expansive 3.0 cm² Imaging Area
Image larger sections or multiple samples on one slide; choose FCX-L (3.0 cm²) or FCX-S (1.25 cm²) and analyze up to ~9.0 cm²/week.
Single-Molecule MERFISH 2.0
Detect RNA at subcellular resolution with high plex and target up to ~1,000 genes per experiment for discovery-scale spatial imaging.
FFPE & Fresh-Frozen Compatible
Run validated workflows that preserve morphology and spatial context across FFPE and fresh-frozen tissue for consistent results.
Integrated Analysis Pipeline
Move from acquisition to insight with Vizualizer (exploration/QC) and VPT (reproducible pipelines) optimized for MERFISH 2.0 outputs.
End-to-End, One Platform
From gene panels to reagents to instrument to analysis. We offer an integrated spatial imaging workflow that speeds setup and keeps teams aligned.
Training, Support & Services
Onboarding, applications help, and lab services when timelines demand it, backed by a global customer-success network.
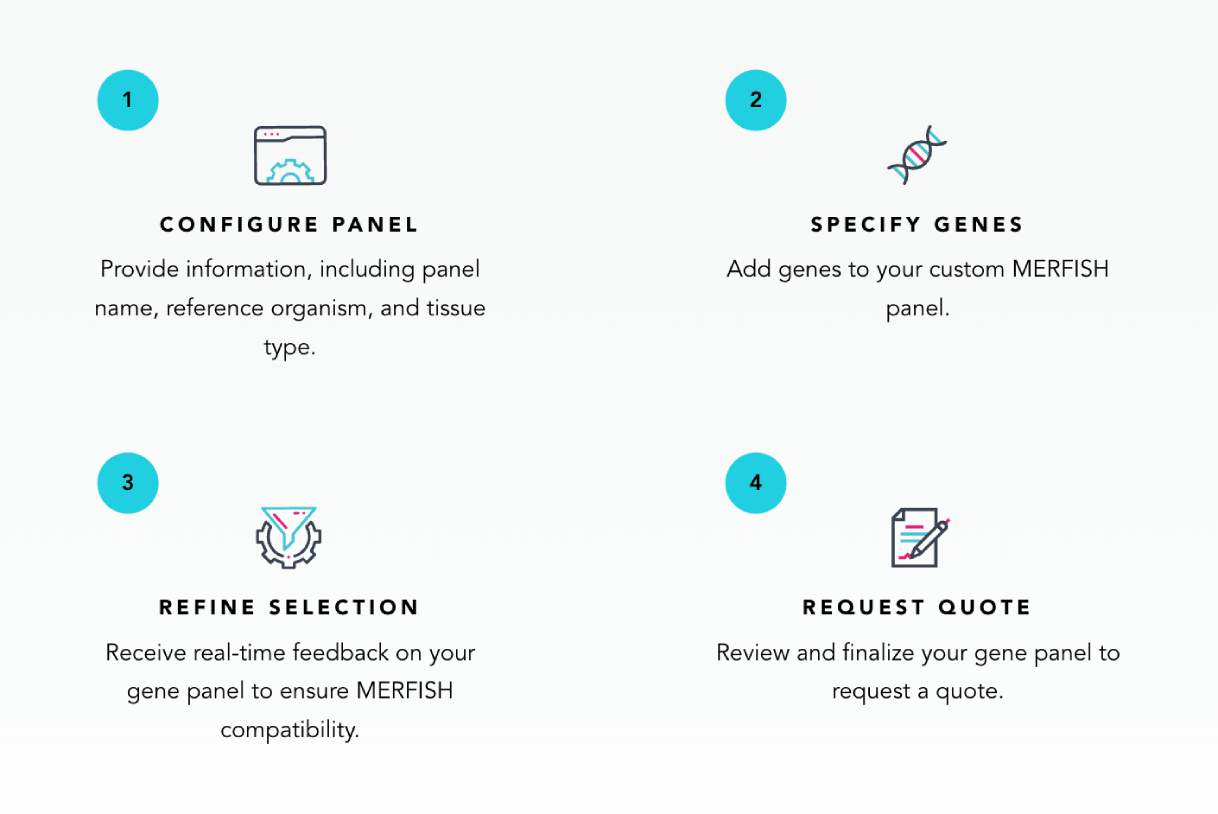
Vizgen Gene Panel Design Portal
The Vizgen® Gene Panel Portal is a user-friendly web application for creating custom gene panels for the MERSCOPE® Ultra Platform.
With the Portal, you can tailor your gene panels with real-time feedback on gene suitability for MERFISH 2.0 measurements. This step-by-step guidance makes it easy to design a panel tailored to your spatial imaging needs.
From the Portal, you can:
- Build custom MERFISH 2.0 gene panels
- Request quotes for gene panels, reagents, and consumables
- Download a panel’s Codebook
- Access previously designed panels and quotes
- Access Vizgen Customer Resources
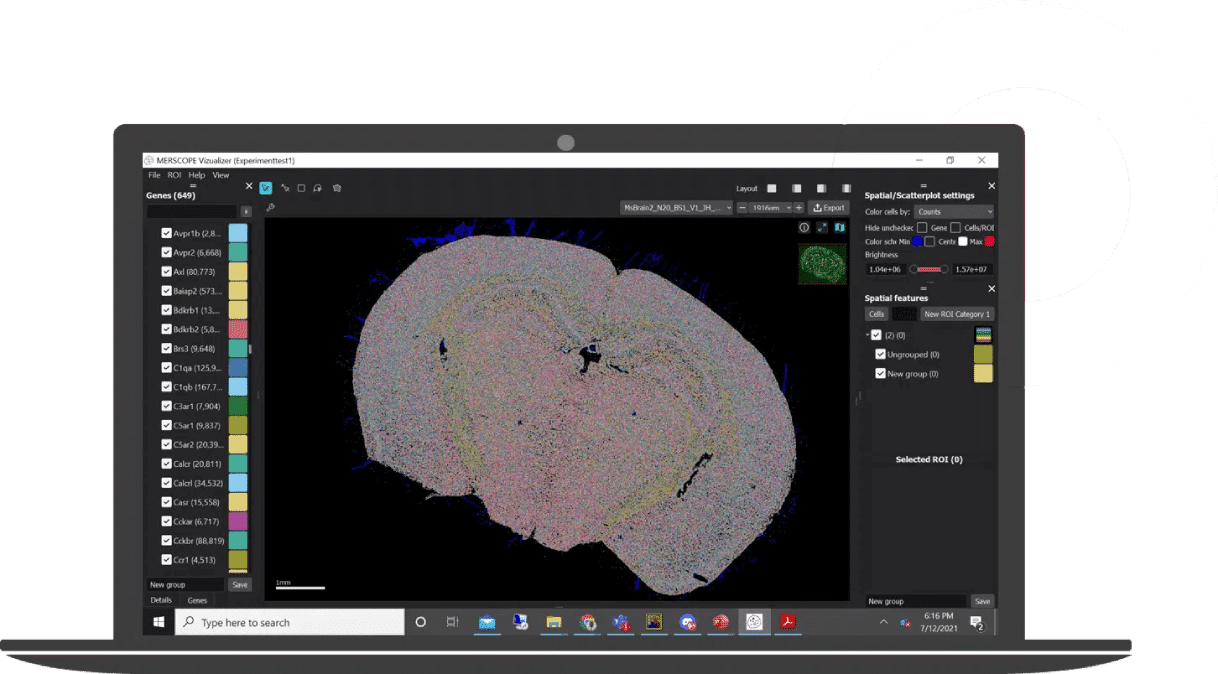
MERSCOPE Vizualizer Software
Explore and interpret complex spatial transcriptomics datasets in Vizualizer™ with intuitive workflows that resolve spatial context at the single-cell level.
With Vizualizer™, you can:
- View tissue images with cell segmentation and boundaries
- Inspect detected transcripts per cell and per gene
- Build cell × gene heatmaps and expression tables
- Explore UMAP/t-SNE scatterplots with cluster annotations
- Export publication-ready figures and share results
Class-Leading Sensitivity
Sub-cellular resolution paired with sample clearing reduces background, ensuring the highest transcript detection.
Platform Flexibility
Custom panels, scalable multiplexing, species versatility, and data analysis tools that work best for your workflow.
High Quality Specificity
Error-robust barcoding and correlation to scRNA-seq provide confidence.
MERFISH 2.0 Chemistry
Enables new applications with high-quality results without sacrificing sensitivity or specificity. More than 170 peer-reviewed journals and pre-prints.
Superior Cell Segmentation
Draw biologically true cell shapes and accurately localize transcripts to identify cell types.
High-Resolution Optics
Minimizes optical crowding and allows for accurate measurements in areas of high expression.
MERSCOPE Ultra Instrument Specifications
Vizgen Flow Chambers Supported
FCX-S (Standard): 1.25 cm² max imageable area; FCX-L (Large): 3 cm² max imageable area
Throughput
Up to 9.0 cm² per week
Multiplex Capacity
Up to 1000 genes
Optical Resolution
Oil-immersion, high-NA objective
Lateral Resolution
100 nm pixel size
Imaging Camera
Back-thinned, cooled sCMOS
On-Instrument Storage Capacity
58 TB
Analysis PC Storage Capacity
58 TB
Illumination Source
Multi-color laser
Automated Image Processing
Transcript decoding and cell segmentation
FAQ
-
What is spatial resolution imaging?
-
How does MERSCOPE™ Ultra achieve single-cell precision in spatial imaging?
-
What types of samples can be analyzed on MERSCOPE™ Ultra?
-
How is MERSCOPE™ Ultra different from other spatial transcriptomics platforms?
-
What is a standard number of targets?
