FFPE Sample Preparation Guide
Get sample prep, imaging, analysis guidance for running MERFISH on FFPE tissue, and practical details to plan cohort-scale immune profiling.
Unlock the complexity of the immune landscape with spatial transcriptomics using MERFISH 2.0™ and proteomics using InSituPlex™ multiplex immunofluorescence (ISP mlF). By combining RNA and protein profiling in situ, immune profiling studies can pinpoint where and how immune cells interact, activate, and influence their surroundings, shedding light on immune regulation in cancer, inflammation, and beyond.
Tumors are heterogeneous and evolve immune-escape strategies, so immune profiling must be done in intact tissue. Using targeted panels, MERSCOPE Ultra can achieve better cell-type resolution, state calling, pathway coverage, and cohort comparability, capturing heterogeneity and immune-escape in intact tissue:
Access FFPE IO datasets built with MERFISH 2.0: curated panels, single-cell maps, and immune annotations that capture heterogeneity in situ.
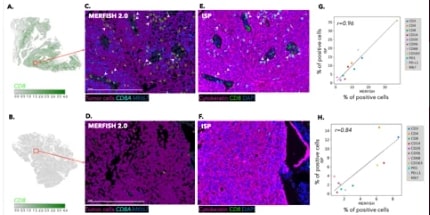
Pairing MERFISH maps with multiplex immunofluorescence on FFPE tissue verifies protein checkpoints and activation states in the same biology, enabling confident immune profiling and proximity analyses.
To see the combined workflow in action, review our AACR posters.
RNA–protein views that explain response vs resistance.
In cancer immunotherapy, response or resistance often emerges from where specific immune states sit and which neighbors they contact. In pre-/post-therapy biopsies (anti-PD-(L)1, CTLA-4, bispecifics, ADCs), MERFISH resolves IFNγ-response programs, TCF7+ progenitors, and M2 macrophage pockets, and relates pathway-score maps (e.g., MAPK, cell-cycle, EMT) to spatial gradients toward or away from tumor nests.
As an example, the Compugen CRC study used MERFISH to refine T-cell subtypes (naïve, Tscm, exhausted; activated vs. quiescent DCs) and discovered PVRIG–PVRL2 interactions at the single-cell level, suggesting a drug strategy to relieve inhibition and restore anti-tumor activity.
When required, matched-region studies pair MERFISH with InSituPlex® multiplex IF on serial sections to align RNA state with checkpoint protein function, strengthening biomarker validation and trial design—and building toward an immune atlas of response mechanisms.

Starting an immuno-oncology study? We’ll help you scope the tissue and endpoints, recommend the right custom or pre-designed gene panels, incorporate matched ISP mIF where valuable, and define analyses for checkpoint expression, spatial proximity, and pathway activity, providing a clear, actionable experimental plan for single-cell immune profiling and developing biomarker-driven trial hypotheses.
Identify T, NK, B, myeloid, and stromal cells and resolve states such as naïve, memory, progenitor-exhausted, plus activated versus quiescent dendritic cells at single-cell resolution.
Quantify checkpoint and activation proteins (PD-1/PD-L1, CTLA-4, LAG-3, TIGIT, Ki-67, GZMB) in FFPE to link RNA programs to protein function in the same tissue regions.
Measure neighborhood enrichment, tumor–immune distances, and ligand–receptor pairs to reveal contact patterns associated with therapeutic response or resistance.
Map chemokines and cytokines (e.g., CXCL9/10/13) and detect tertiary lymphoid structures to characterize immune niches and infiltration barriers.
Generate in-tissue pathway scores (MAPK, cell cycle, IFNγ, EMT, senescence) per population to connect spatial context with mechanism, advancing single-cell immune profiling.
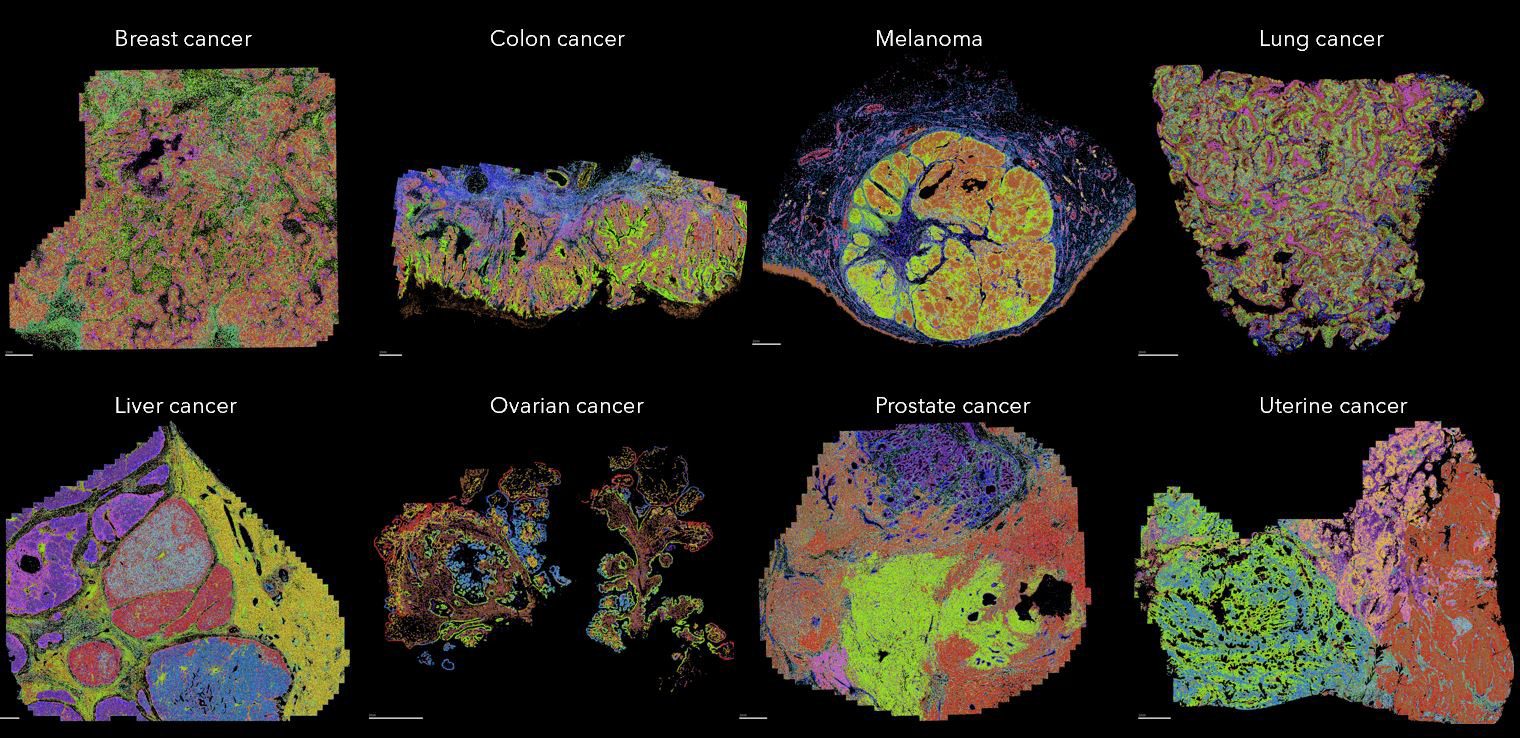
Figure: MERSCOPE Platform maps the spatial location of different cell types across multiple FFPE human cancer samples.
Spatial profiling using MERFISH 2.0 resolves exhausted and progenitor-exhausted T-cell programs and, when paired with multiplex IF, quantifies PD-1/PD-L1 and other checkpoint phenotypes in situ, enabling responder stratification based on cell state and proximity.
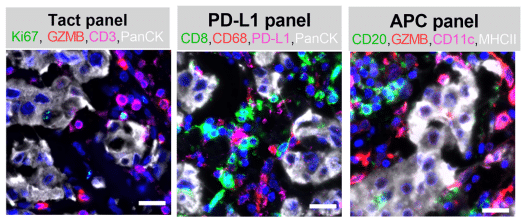
Figure: Human 4-plex panels: concordance assay. (A) Shows representative images illustrating the immunolabeling pattern of Ultivue Tact, PD-L1, and APC panels in human breast tumor resections.
Across FFPE cohorts, MERFISH builds an immune cell atlas by subtyping T, NK, B, and myeloid populations, mapping niches and tertiary lymphoid structures, and comparing tissue sites to reveal indication-specific patterns.

Figure: Spatial distribution of select genes within a magnified region in human breast cancer, with CD4 (green), CD8A (blue), FOXP3 (red), NCR1 (yellow), and CTLA4 (white) shown. Note that FOXP3-positive Tregs express T cell exhaustion marker CTLA4, marked by the red arrowheads. NK cell marked by the yellow arrowheads.
Standardized FFPE workflows and predesigned panels deliver consistent single-cell data across many slides and sites, enabling cohort studies with reproducible analyses for translational immune profiling.

Figure: Murine 4-plex panel: reproducibility assay. Shows representative fluorescence images of the biomarkers across
the different runs. Scale bar equals 50 microns.
Designing a cancer gene panel that truly reflects disease biology takes time and careful curation. MERFISH® 2.0 enables high-plex spatial transcriptomics, while InSituPlex® provides pathology-grade spatial proteomics. Used as complementary workflows, these approaches build a more complete picture of tumor, immune, and stromal interactions across the microenvironment.
Peer-reviewed studies show how Vizgen MERFISH and Ultivue multiplex immunofluorescence enable rigorous, FFPE-ready immune profiling: validated, quantitative protein maps; whole-slide PD-L1 and immune-cell heterogeneity across primary and metastatic tumors; and MERFISH-defined DC–T helper niches linked to CD8⁺ T-cell differentiation and PD-1 response.