Spatial Biology Platforms
Discover two complementary spatial biology platforms: MERFISH® for spatially resolved transcriptomics and InSituPlex® for multiplex immunofluorescence proteomics, enabling unprecedented multiomics insight.

MERFISH 2.0: Spatially Resolved Transcriptomics
Single-molecule FISH for high-plex, single-cell RNA maps with subcellular precision.
Measure the copy number and spatial position of thousands of RNA species in intact tissue using combinatorial barcodes and iterative imaging. Optimized probe chemistry boosts sensitivity and specificity, enabling robust data even in challenging FFPE samples.
These capabilities make MERFISH 2.0 a cornerstone of Vizgen’s spatial biology platforms, revealing cell states, interactions, and tissue architecture at single-cell resolution.
Our Services

Spatial Transcriptomics
Explore Vizgen’s spatial transcriptomics analysis services using MERFISH 2.0™ on the MERSCOPE Ultra™ platform. Our team delivers single-cell, spatially resolved gene-expression data across tissue types with custom panel design and high-precision imaging.
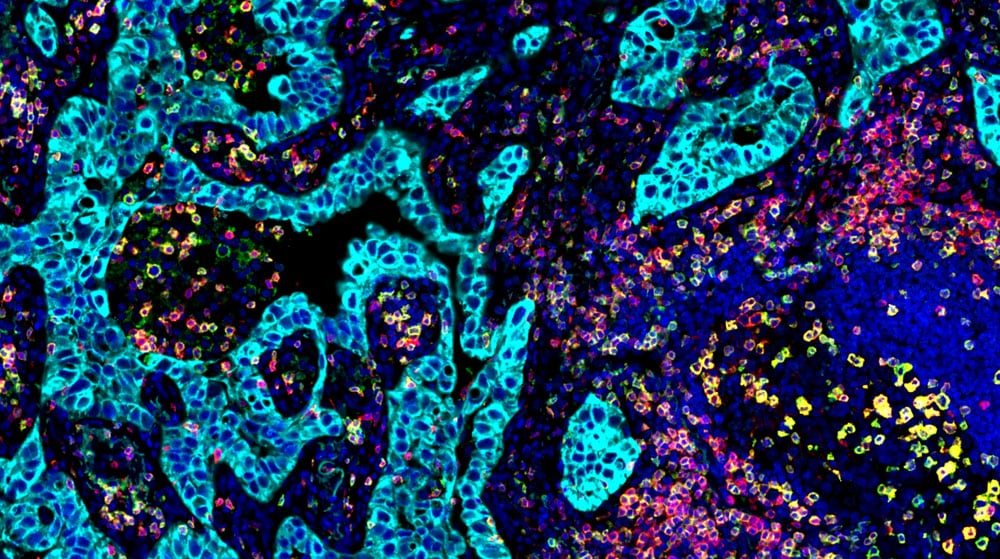
Spatial Proteomics
Advance discovery with Vizgen’s assay development services powered by InSituPlex® multiplex immunofluorescence. From antibody selection and validation to imaging and STARVUE™ analysis, we provide end-to-end expertise for accurate, multiplex protein detection.
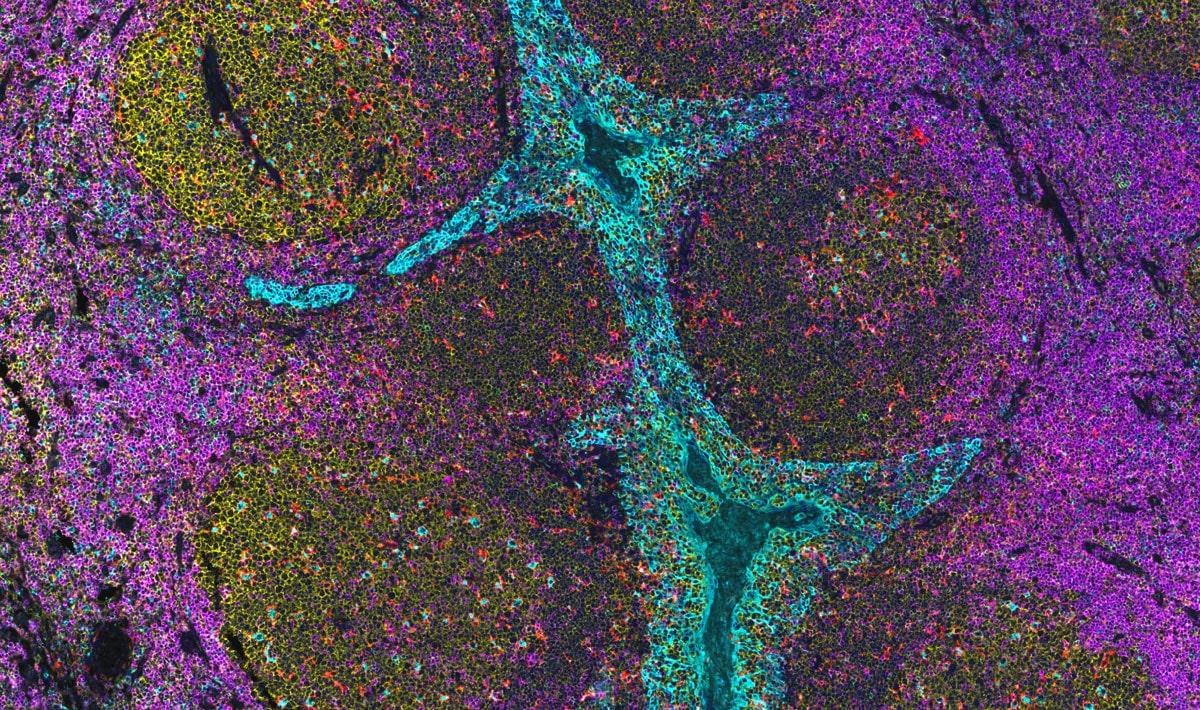
InSituPlex: Multiplex Immunofluorescence
DNA-barcoded IHC that delivers quantitative, pathology-grade protein detection.
Deploy DNA-barcoded primary antibodies for direct detection, then amplify the signal in proportion to antigen abundance before sequential imaging rounds decode many markers in the same section. Achieve quantitative, high-specificity, wide-dynamic-range protein maps that preserve morphology and scale across FFPE and frozen tissue.
As a pillar of Vizgen’s spatial biology platforms, InSituPlex complements MERFISH 2.0 by localizing proteins and activity.
Single-Cell Spatial Resolution
Map RNA and protein expression at true cellular resolution to reveal biological context in intact tissue.
Scalable Multiplex Detection
Profile hundreds to thousands of targets in the same sample for deep, unbiased molecular insights.
Integrated Spatial Biology Platform
Combine transcriptomic and proteomic data for a complete picture of biological systems.
