A Special Issue of Science Featuring BICAN Research
According to Science, the human brain is composed of 86 billion neurons and a similar number of non-neuronal cells. Over the last decade, there has been a global push to better understand the complex cellular composition of the human brain. A recent special issue of Science put a spotlight on a collection of research papers that set out to uncover the genetic organization of the adult and developing human and nonhuman primate (NHP) brain, building a collective comprehensive brain cell atlas.
An exciting five of the twenty-one studies featured in the special issue leveraged Vizgen’s MERSCOPE® Platform to complete their research, reaffirming MERSCOPE’s position as the industry’s leading spatial genomics solution.

Cover of Science: Brain Cell Census VOLUME 382|ISSUE 6667|13 OCT 2023. Image illustration depicts the left hemisphere of the human brain, with colored pixels representing the cell types composing the brain.
The five MERSCOPE studies investigated the composition, structure, and spatial organization of cells in various regions of the adult and developing human and NHP brain, with the results answering key questions about what makes our brains human, how the cellular makeup of the brain has changed throughout human evolution and changes with age, and how we can predict vulnerabilities to developmental brain disorders.
Tomasz Nowakowski, a University of California, San Francisco neuroscientist and an author of two of the MERSCOPE studies, shared his excitement in a Wall Street Journal article on the data that are enabling researchers to better understand the human brain and its genetic organization. “It’s a dream that has been around for more than a century. This is like building a map of the universe,” said Nowakowski in the article.
Powered by MERFISH spatial transcriptomics technology, MERSCOPE enables researchers to directly map, quantify, and characterize gene expression across intact tissues down to subcellular resolution, revolutionizing life sciences research.
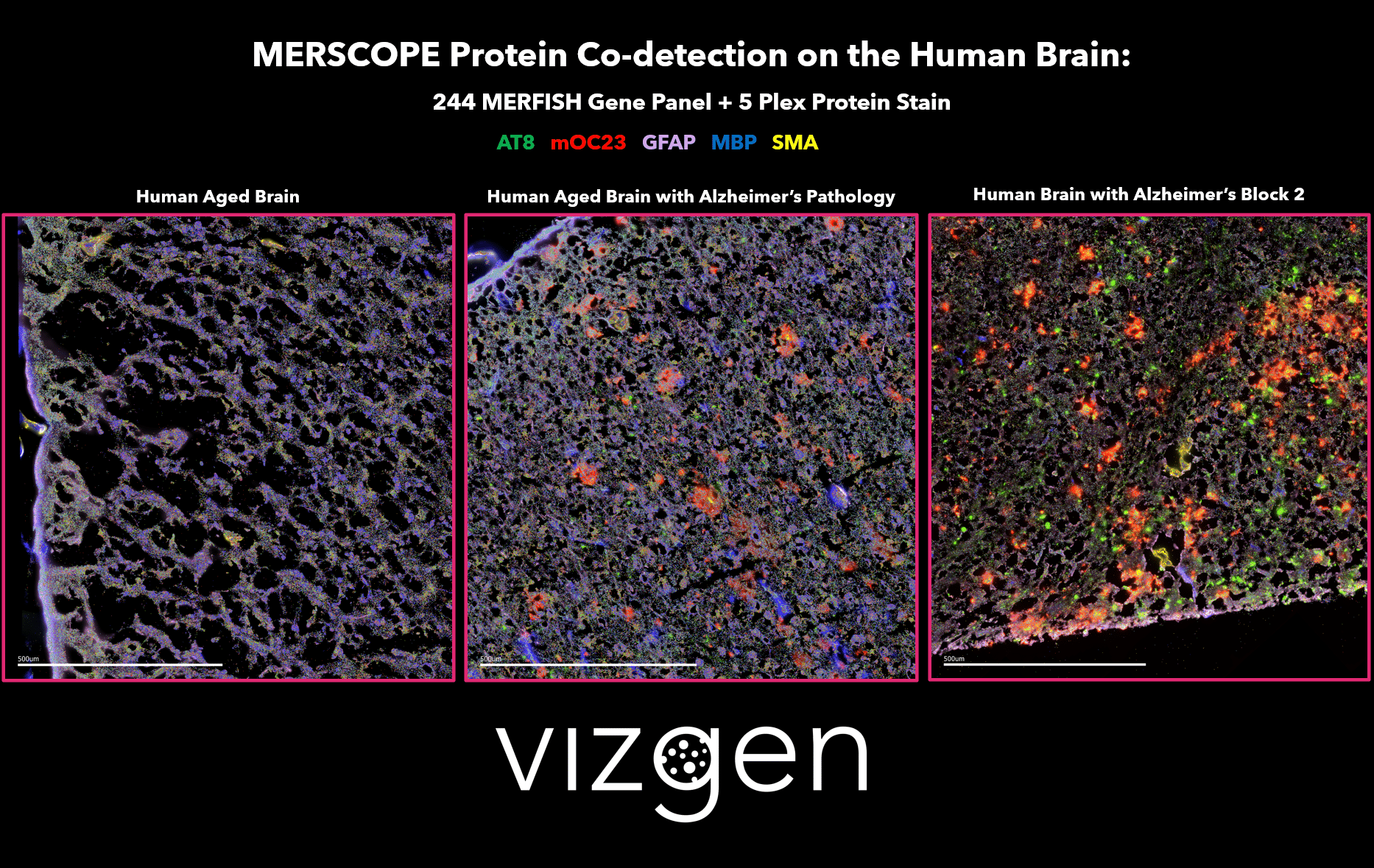
FIGURE 1: MERSCOPE Data Image showing the spatial multiomic differences between a normal human aged brain, and human brains with Alzheimer’s disease. The data image shows the spatial distribution of detected transcripts from 244 genes and 5 co-detected proteins across intact human brain tissue samples.
MERSCOPE studies contributing to the human brain cell atlas
Transcriptomic cytoarchitecture reveals principles of human neocortex organization
Rebecca Hodge, Ed Lein, and Trygve Bekken’s group (Jorstad et al., Science) elegantly defined quantitative cellular cytoarchitecture across a series of neocortical areas using single-nucleus and MERFISH spatial transcriptomics. Researchers observed relative proportions of neuronal subclasses as a fraction of all excitatory or inhibitory neurons in each neocortical area and estimated based on snRNA-seq profiling or in situ labeling using MERFISH. Based on MERFISH in situ labeling experiments, laminar distributions of L2/3/4 IT and SST types were also identified, revealing principles of human neocortex organization.
Interindividual variation in human cortical cell type abundance and expression
Rebecca Hodge and Trygve Bekken’s team (Jorstad et al., Science) were able to identify human-specific cortical features using single-nucleus and MERFISH spatial transcriptomics. The team leveraged a 140-gene MERFISH panel designed to identify human cortical cell types to map every type described in an updated human middle temporal gyrus (MTG) taxonomy to determine cell-type locations within the cortex and confirm cell-type proportions. The MERFISH data also validated the subtypes within L5/6 IT Car3 neurons based on the expression patterns of CUX2 in human and macaque MTG, characterizing key differences in the cortical features of the human and macaque brain.
Single-cell analysis of prenatal and postnatal human cortical development
Arnold Kriegstein’s group (Velmeshev et al., Science) identified cortical cell types and lineages across brain development using single-nucleus and the MERSCOPE Platform. The authors used 140 lineage-specific genes to visualize the spatial map of annotated cell types across prenatal and postnatal human cortical development, with their results shedding light on lineage-specific mechanisms of normal cortical development.
Spatiotemporal molecular dynamics of the developing human thalamus
Tomasz Nowakowski’s team (Kim et al., Science) generated a reference map of the developing human thalamus and validated the cell types using MERFISH spatial transcriptomics. Researchers were able to map the regions, cell types, and selected marker genes of the first-trimester thalamus by using MERFISH. Maps showcase the spatial clustering analysis of MERFISH data of the thalamic glutamatergic neuron subtypes in the human mid-gestation thalamus, uncovering spatiotemporal molecular dynamics of the developing human thalamus.
Signature morphoelectric properties of diverse GABAergic interneurons in the human neocortex
Ed Lein and Jonathan Ting’s groups (Lee & Dalley et al., Science) utilized Patch-seq and MERFISH to gain deeper insights into morphoelectric properties of GABAergic interneurons in the human neocortex. The authors first used Patch-seq to characterize the electrophysiological, morphological, and transcriptomic profiles of GABAergic neurons in human neocortical brain slices. Using MERSCOPE, the authors were then able to map the spatial distribution of all types in the MTG taxonomy, showing that the transcriptomic organization was in close alignment with soma distributions from Patch-seq experiments.
Our team is proud that the MERSCOPE Platform has played an integral role in assisting these researchers with creating a comprehensive human brain cell atlas and expanding the understanding of how our brains have developed and work. We look forward to MERSCOPE continuing to be a driving force in transformative discoveries in the field of neuroscience and beyond.
Contact us to learn more about how MERSCOPE can help to advance your research or get in touch with our Lab Services team for a project consultation.
Visit the Vizgen Data Release Program database to explore the MERFISH Mouse Brain Receptor Map and experience MERSCOPE spatial data firsthand!